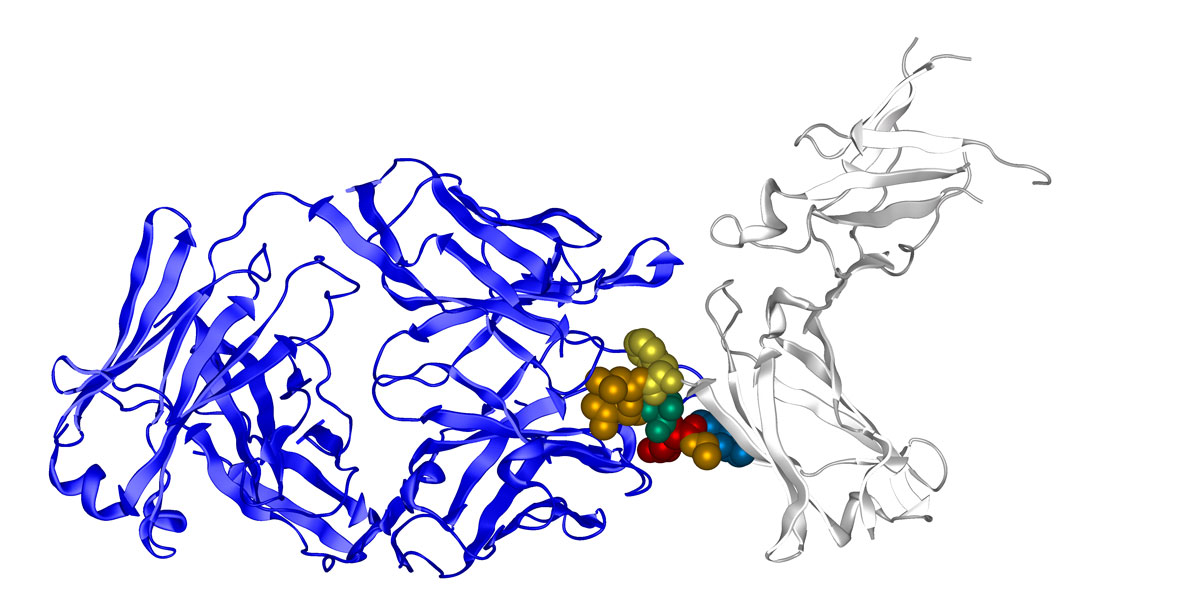
Epitope fingerprints reveals the amino acids essential for binding and, when required for binding, additional structural constraints. The information is obtained from a single experiment, and it truly represents an antibody’s individual epitope fingerprint. Ideally, the entire procedure takes only 2-3 weeks and requires minimal amounts of IgG antibody, even with unpurified protein samples.
Because all peptides selected are expressed on phage particles, the identified disulfide bridges fold properly, even in a synthetic peptide.
Epitopes can be identified even for antibodies against unknown antigens. If a sufficiently large number of amino acid residues are enriched, antigen identification can be performed using protein database searches.
In most of the currently applied standard epitope mapping experiments, an epitope is often approached by roughly describing the region or sequence of the antigen recognized by an antibody. Compared to the epitopic approach, these methods require much more time and materials for peptide screening, alanine scanning, structural optimization, and positional screening of alternative amino acids.
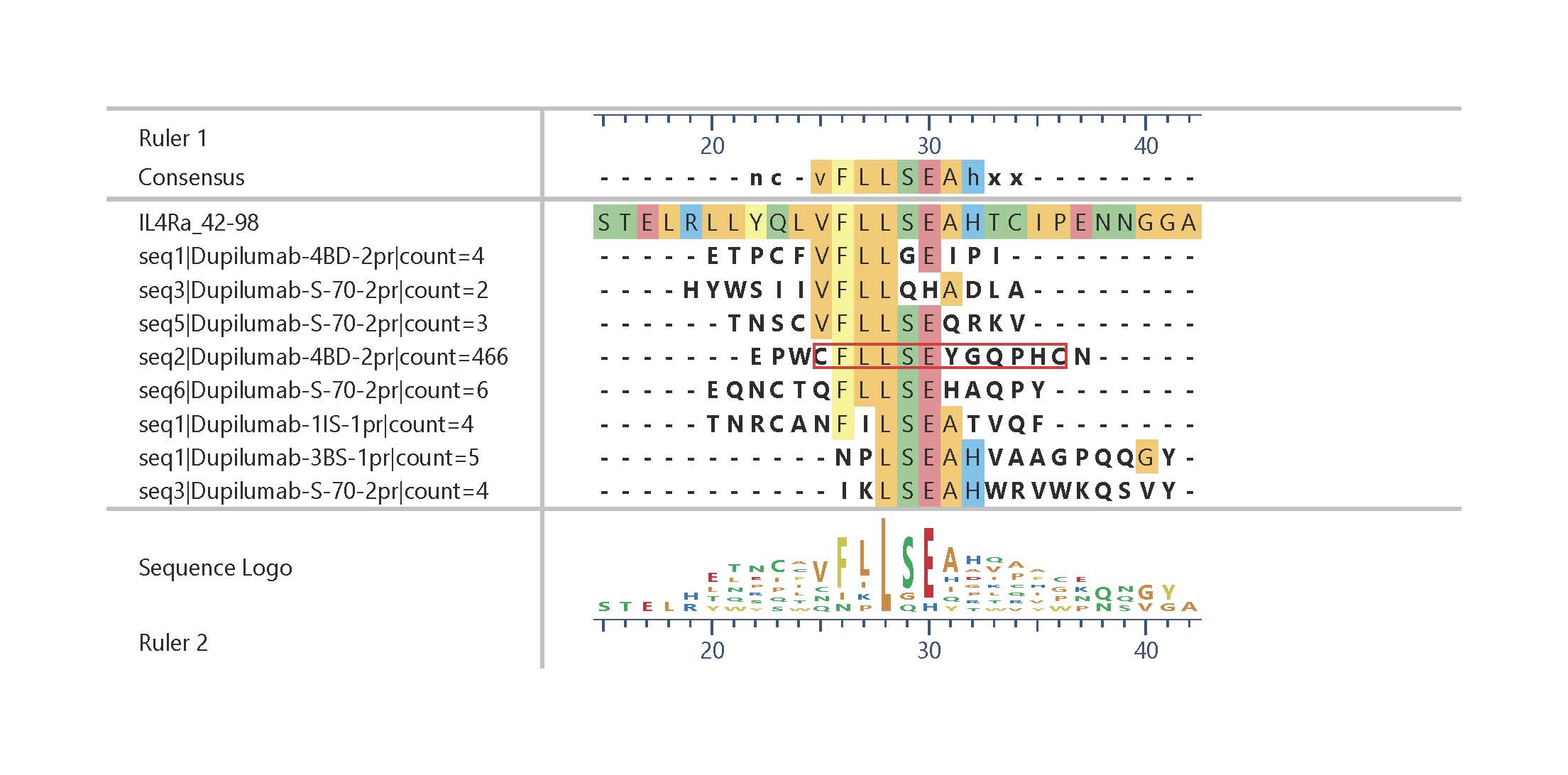
Epitope fingerprint of Dupilumab identified by epitope fingerprinting. The alignment shows only sequences with five consecutive identical amino acids out of a pool of more than 2,000 sequences with at least 4 amino acid identities. Strongest enrichment: Cys-flanked constrained peptide (red frame). Click on this link or the graphic to obtain a longer list of similar sequences.