EPITOPIC’s approach, searching for peptides binding to antibodies or other targets, is unique in saving time and materials by reducing wet-lab procedures to a minimum and applying a statistical approach to analyze large datasets of peptide sequences obtained using NGS.
The procedure begins by capturing binders from a proprietary random library of 5×109 16-mer peptides. This naïve library can be used for all projects. It has the unique feature of well-controlled distribution of individual amino acids. Small deviations from the statistically expected frequency are detected using a specially developed software package. Indeed, a few hundred antibody molecules in the selection experiment are sufficient to identify pools of binding sequences in hundreds of thousands of sequences obtained by NGS. Please refer to Epitope Fingerprinting Process for further details
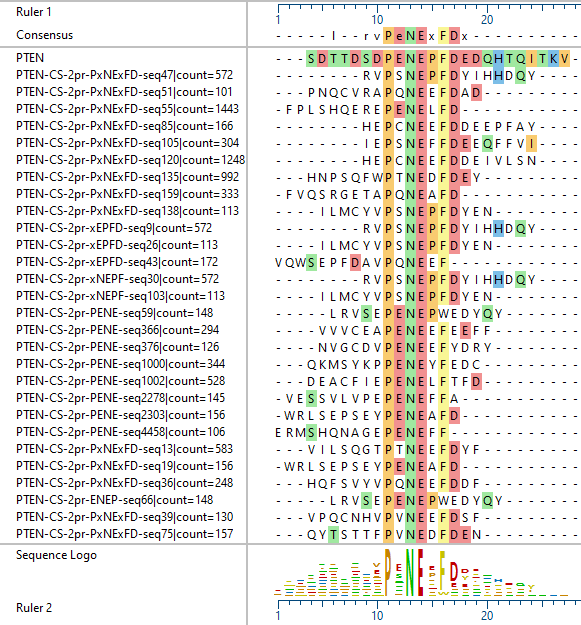
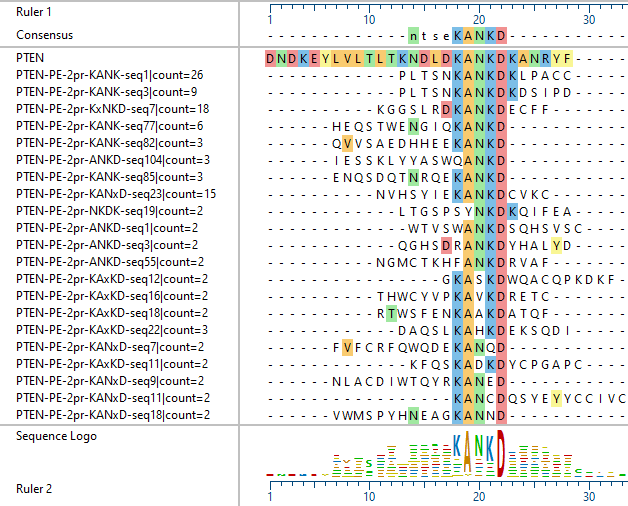
The library covers almost all possible 6-mer sequences, including discontinuous variants; therefore, aligning these candidate binders allows prediction, for example, not only of an antibody epitope. It also tolerated amino acid variations and conserved adjacent residues, helping to understand binding to discontinuous epitopes. In contrast to approaches using simple linear peptides or libraries of peptides derived from the antigen, the library structure allows the selection of structurally constrained peptides without applying special libraries or time-consuming synthesis of peptides.
The variety of epitopes identified is reflected in the following examples of different antibodies against the PTEN protein: