Epitope Fingerprinting with polyclonal Antibodies
A few hundred antibody molecules are sufficient for epitope mapping. A single selection experiment with 20-100 µL serum yields enough data to cover several hundred epitopes! This application has been successfully tested and optimized in many epitope fingerprinting projects. Because the sensitivity of this approach is high and the number of antibody molecules required for successful epitope fingerprinting is low, even very diluted polyclonal antibody solutions can be mapped. Once peptide datasets are generated, they can be searched repeatedly for antibody epitopes in any suspected antigen. This is the perfect way to save time and money.
It is important to remember that we use a completely naïve approach instead of dedicated libraries. Epitope identification is based solely on the statistical analysis of NGS data comprising several hundred thousand peptide sequences. This allows the identification of binding peptide motifs to hundreds of antibodies, including most allowed variations of the peptides and structures potentially binding the antibody, and to predict species and cross reactivity for individual antibodies. This has recently been published for food allergies.
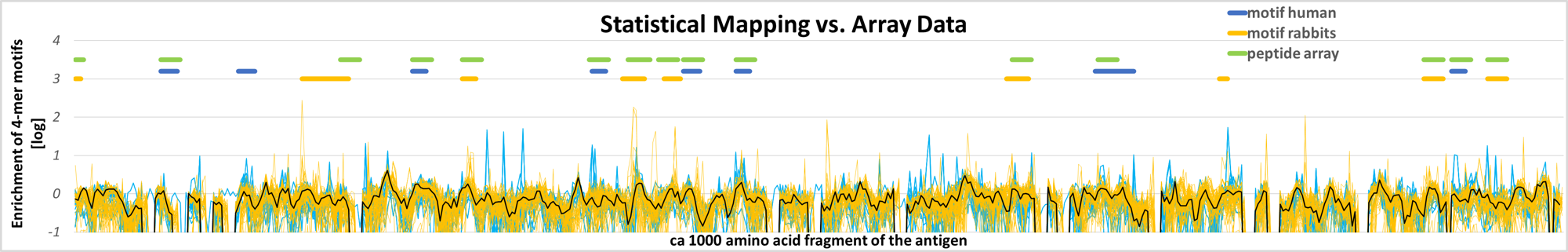
Mapping of vaccine and infectious agent epitopes in sera from vaccinated rabbits, infected patients, and infected or vaccinated pigs. Sites for epitopes from selections on these sera are visible as enrichment of 4-mers comprising the antigen protein. The identified peptide epitopes have been validated in peptide arrays, and the immune responses of different species are surprisingly similar.